DNA Fingerprinting
Historical background
DNA fingerprinting was developed in 1984 by Alec. J. Jeffrey at the University of Leicester while he was studying the gene for myoglobin.
He found that myoglobin genes contain many segments that vary in size and composition and have no apparent function.
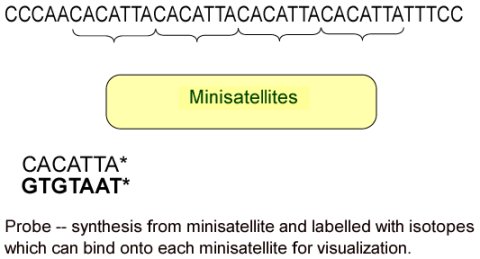
Jeffrey called these segments minisatellites because they were small and surrounding the part of the gene.
The minisatellites account for less than 1 percent of the total DNA of human.
DNA Probes
Jeffrey isolated several of these minisatellite genes and inserted one of them into bacteria.
Large amount of the DNA segments could then be produced and purified.
Label the DNA segments with radioactive isotopes to produce DNA probes.
What is DNA Fingerprinting used for
At present, DNA fingerprinting is the most powerful and authoritative method to identify an individual due to the different polymorphisms.
With the exception of identical twins, the genetic make-up of every human being is different from one another.
There are two types of DNA structural polymorphisms:
1. Site polymorphism - change of one or more nucleotides at a particular site
2. Length polymorphism - number of repeating units is different at a particular locus.
Common Types of DNA Fingerprinting
Restriction Fragment Length Polymorphism (RFLP)
An example of site polymorphism :
1. Isoloation of DNA
a. DNA samples are obtained from substances such as blood, semen, hair roots, or saliva. b. The individual cells from the sample are split open, and the DNA is separated from the rest of the cellular debris. (DNA extraction) c. Using polymerase chain (PCR) to multiply the amount of DNA. 2. Cutting with restriction enzyme
a. The DNA is then treated with restriction enzymes which cleave the DNA into smaller fragments by cutting at specific sites. b. The result is a unique collection of different-sized fragments. c. Since the minisatellites from any two individuals have different compositions, they are cleaved at different sites, producing fragments of different lengths. d. These different lengths of fragments existing between individuals are called as restriction fragment length polymorphisms (RFLPs) 3. Gel Electrophoresis a. The DNA fragments are then applied to one end of a thin, jellylike substance called an agarose gel. b. An electric current is passed through the gel for a short period of time. c. The negatively charged DNA fragments will migrate across the surface of the gel in response to the current. d. The smaller and more mobile pieces of fragments will travel farther. e. The DNA is thus separated into different bands with fragments in each one progressively getting smaller in size. 4. Blotting
a. Because the gel cannot be easily handled, a thin nylon membrane is laid over its surface and covered by layers of paper towels. b. As the towels draw moisture from gel, the DNA is transferred onto the surface of the nylon membrane. This process is called blotting. c. Soak the nylon membrane overnight. 5. Probing
a. The DNA bands are still invisible to the eye, and there are too many to be useful. b. Therefore, a solution of the radioactive probes made from minisatellites is washed over the surface of the membrane. c. If any of the probes have the same compositions as a part of a DNA fragment, they will bind together. d. The probes will ignore the vast majority of the hundreds of bands present. 6. DNA fingerprint
a. To see the pattern of bands, place a sheet of photographic film on top of the membrane. b. The radioactive labels will expose the film and ultimately produce a pattern of thick and thin dark bands. c. This pattern of bars is the DNA fingerprints.
Common Types of DNA Fingerprinting
Short Tandem Repeat (STR) Analysis
An example of length polymorphism
1. Isolate DNA
2. Copy a specific fragment many times (amplification) using the polymerase chain reaction (PCR)
3. Separate the amplification products according to the size by electrophoresis
4. Compare the size pattern of the amplification products
5. The pattern obtained may vary between individuals because of the length polymorphism
Applications of DNA Fingerprinting
Human Identification
- Crime Investigation
- Population DatabaseHuman Relation
- Paternity Test (who is the biological father)
- Maternity Test (who is the biological mother)
- Grandparentage
- Sibling-ship (brothers/sisters)
- Deceased PersonIdentification of Genetically Modified Plants & Animals
Population Studies, e.g. Migration of Ethnic Groups
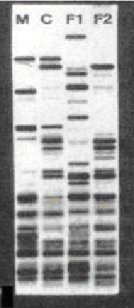
Case Study 1: Who is the Father?
The DNA bands of Child, C, can be found either in mother, M, or in F2; but not in F1.
Therefore F2 is the biological father of the Child.
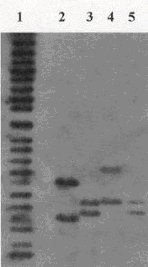
Case Study 2: Who Commits the Crime?
Lane 1 = molecular weight standard
Lane 2 = victim
Lane 3 = evidence
Lane 4 = suspect 1
Lane 5 = suspect 2As shown, the DNA pattern of evidence is identical to the DNA pattern of suspect 2.
Therefore suspect 2 is the criminal.
Reference
1. DNA Fingerprinting Services
(http://www.geneprolab.com)
2. Frequently asked questions about paternity testing (http://www.geneticid.com/we01001.htm)
3. How does a murderer traced through blood samples? (http://www.faseb.org/genetics/gsa/careers/bro-05.htm)
4. DNA in the courtroom
(http://library.thinkquest.org/19037/court.html)
Copyright(c)2002 HKIEd. All Rights Reserved.
請採用 Internet Explorer 4.0 或 Netscape Navigator 4.0 或以上瀏覽器,並以800x600解析度瀏覽。